Translational regulation in bacterial pathogens
The MtbTransReg Project:
Mycobacterium tuberculosis is the causative agent of human tuberculosis (TB), one of the humankind’s deadliest diseases. The World Health Organisation estimated that there were 6.3 million new cases of TB and 1.3 million TB deaths in 2016. Furthermore, it is estimated that one-third of the world’s population harbours the bacterium in the form of an asymptomatic infection referred to as latent TB. This reflects the complex life cycle of the bacteria that can involve prolonged periods of non-replicating persistence prior to the active disease process that is required for onward transmission. During this process, the bacteria overcomes numerous stresses presented by the immune system of the host. Still today, the mechanisms underlying persistence are poorly understood, and the emergence of drug-resistant bacteria makes the development of effective new treatments an urgent challenge. Understanding the ability of M. tuberculosis to switch between replicating and non-replicating states during infection and disease is central to the search of improved treatments.
Translation, the process by which the sequence of nucleotides in a gene directs the synthesis of proteins, is an intricate process involving many cellular components. Amongst these, the ribosome has 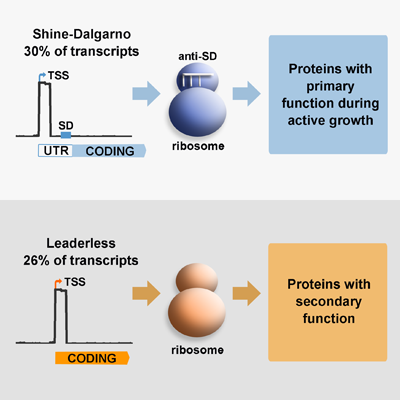 been traditionally considered as a conserved nucleoprotein with the only role of mediating this process. Genes contain specific signals that optimise their interaction with ribosomes, known as leader sequences, these include the Shine-Dalgarno (SD) sequence required for canonical translation initiation in bacteria. There is recent evidence that suggests that ‘specialised ribosomes’, these are modified ribosomal particles, can modify the proteome profile by preferential translation of particular gene subsets, particularly in response to stress. M. tuberculosis differs from other important human pathogens in expressing a large number of leaderless genes, which do not have the SD sequence required for canonical translation initiation. In the model bacteria Escherichia coli, only a few leaderless genes have been described and they are selectively translated by specialised ribosomes upon stress conditions. We have previously demonstrated in M. tuberculosis, that under conditions of nutrient starvation the abundance of leaderless genes increases, suggesting that translation of leaderless genes may be an important component of the adaptive response of this pathogen.
been traditionally considered as a conserved nucleoprotein with the only role of mediating this process. Genes contain specific signals that optimise their interaction with ribosomes, known as leader sequences, these include the Shine-Dalgarno (SD) sequence required for canonical translation initiation in bacteria. There is recent evidence that suggests that ‘specialised ribosomes’, these are modified ribosomal particles, can modify the proteome profile by preferential translation of particular gene subsets, particularly in response to stress. M. tuberculosis differs from other important human pathogens in expressing a large number of leaderless genes, which do not have the SD sequence required for canonical translation initiation. In the model bacteria Escherichia coli, only a few leaderless genes have been described and they are selectively translated by specialised ribosomes upon stress conditions. We have previously demonstrated in M. tuberculosis, that under conditions of nutrient starvation the abundance of leaderless genes increases, suggesting that translation of leaderless genes may be an important component of the adaptive response of this pathogen.
The MtbTransReg project aims at understanding what is the role of selective translation of leaderless and SD genes in the context of adaptation to stress and drug resistance in M. tuberculosis. We are tackling these questions by using cutting-edge experimental techniques combined with bioinformatics analyses.
The MtbTransReg project is funded by the European Research Council and you can find more information about it here
