Cell Lines & Plasmids
2T1 ‘landing pad‘ bloodstream-form (BSF) T. b. brucei cell line derived from strain 427/MITat1.2 (Alsford et al Mol Biochem Parasitol 144:142-8) was developed to overcome the rDNA spacer ‘position effect’. This ensures robust and consistent tetracycline-inducible expression. The cell line expresses TetR from the pHD1313 cassette integrated at the tubulin locus (see Alibu et al Mol Biochem Parasitol 139: 75-82).
Some useful protocols: culture of BSF T. b. brucei; electroporation and selection of transformants
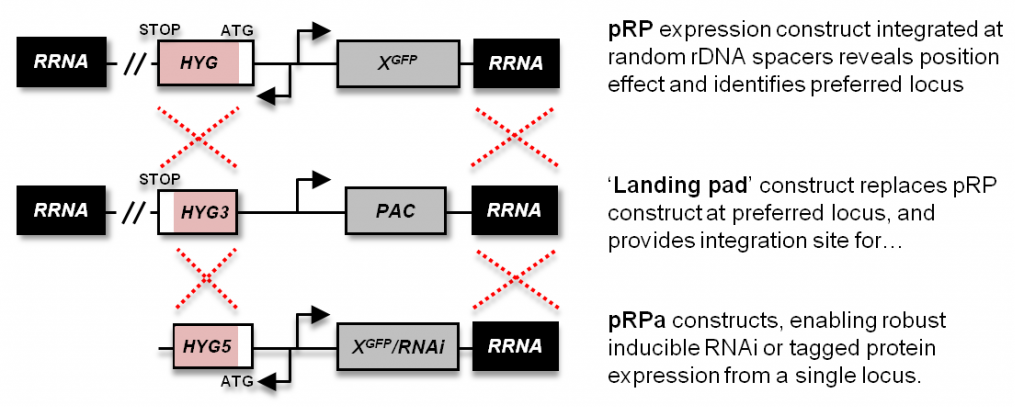
The ‘landing pad‘ is integrated at a non-transcribed rDNA spacer capable of supporting consistent high level RNA polymerase-I transcription. This recombinant locus consists of a 3′-HYG fragment and VSG expression site promoter-driven PAC ORF. Integration of a pRPa plasmid at this locus reconstitutes the HYG ORF and deletes the PAC ORF, rendering transformants resistant to hygromycin and sensitive to puromycin.
Prior to transfection, 2T1 (VSG221 expressing, Tagged, clone 1) T. b. brucei should be maintained in 1 µg/ml phleomycin (TetR) and and 1 µg/ml puromycin (‘landing pad‘).
pRP plasmid series | sequences | maps | details
A series of constructs that integrate at a random rDNA spacer. Transformants exhibit a range of insert expression levels depending on the rDNA spacer at which they integrate. Versions are available for MYC and GFP tagging (BLA or HYG selectable markers), and stem-loop RNAi (HYG). These can be used in any T. brucei cell line that isn’t already resistant to hygromycin or blasticidin.
ph3Ep – ‘landing pad‘ plasmid | sequence
This plasmid consists of a 3′-HYG fragment and a VSG ES promoter-driven PAC ORF. It replaces an integrated pRP(HYG) expression construct, generating a landing pad locus that can be reproducibly targeted by the pRPa plasmids (see below), and cells that are hygromycin sensitive and puromycin resistant.
For more details, see Alsford et al (2005) Mol Biochem Parasitol 144: 142-8.
pRPa plasmid series | sequences | maps | details
A series of constructs that integrate at a rDNA spacer containing an integrated landing pad; the pRPa plasmids can only be used in the 2T1 landing pad cell line. These plasmids differ from the pRP series in two ways: they carry a 5′-HYG fragment and have AscI sites flanking the integration sequences (for linearisation prior to integration at the landing pad locus). Versions are available for MYC and GFP tagging, and stem-loop RNAi. We also have a limited number of constitutive constructs that lack the Tet operator.
pRPa transformants exhibit consistent cassette expression levels, as integration always occurs at the landing pad locus (all derived lines are hygromycin resistant). However, not all lines become puromycin sensitive, suggesting that cross-over sometimes occurs upstream of the landing pad VSG ES promoter. If you require your derived cell lines to be puromycin sensitive to enable downstream manipulations, you’ll need to screen your transformants – normally >50% of transformants are puromycin sensitive, though it can be less for those transformed with a stem-loop RNAi pRPa construct.
pNAT construct series
N-terminal tagging | maps | details
C-terminal tagging | maps | details
A series of GFP and multi-MYC constructs for tagging endogenous loci at the N- or C-termini. Variants carrying BLA and HYG selectable markers are available. These can be used in any T. brucei cell line that isn’t already resistant to blasticidin or hygromycin.
For more details, see Alsford & Horn (2008) Mol Biochem Parasitol 161: 76-9.
Details of additional T7 polymerase-driven tagging and RNAi plasmids are available on the Horn Lab Resources page.
If you would like to use the 2T1 cell line or any of these plasmids in your work, please feel free to contact myself or David Horn.
The 2T1 cell line and the constructs described above have been widely used by ourselves and others; details can be found here (2T1) and here (constructs).
If you’ve used the 2T1 cell line or these plasmids, and have any comments or questions about them, please feel free to get in touch. Also, if you’ve modified any of the plasmids with your own preferred fluorescent or epitope tags, or cloning system, please let me know; this will help me to maintain a comprehensive list of available plasmids, enabling others in the field to benefit from them.